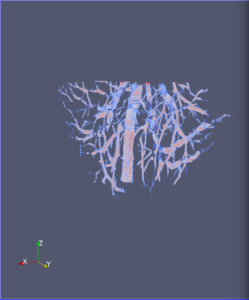
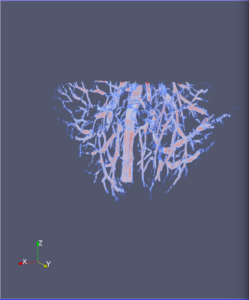
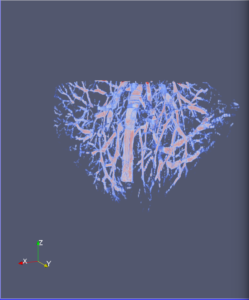
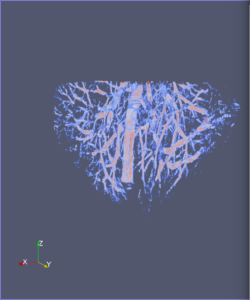
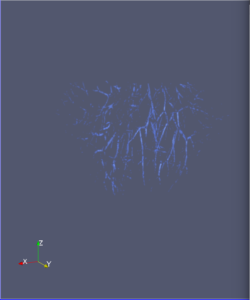
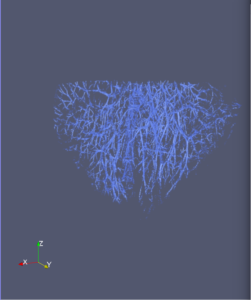
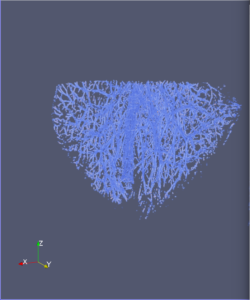
Computational 2D and 3D extraction of liver mouse vasculature within synchrotron radiation X-ray phase tomography
Hugo Rositi, Aurélie Leborgne, Marie-Ange Lebre, Manuel Grand-Brochier, Fabien Chauveau, Marlène Wiart, Cécile Olivier, Françoise Peyrin, Antoine Vacavant
Abstract:
Extraction and characterization of liver vascular network is of high interest in several computer-aided biomedical applications such as angiogenesis prediction, tumor’s blood distribution, numerical simulation of blood flow and embolization [1], etc. At a macroscopic scale, different imaging modalities give access to vessel internal information and vascular trees through 3D imaging as MRI or CT scan for instance, and 2D acquisitions, e.g. angiography. In these modalities, the finest vessels cannot be visualized due to the constraints coming from system resolution (approximately 1 x 1 x 1 mm3 in an anisotropic way). For small animals and to get into a more precise study, angiography at a microscopic scale is needed and still remains an open challenge even if several approaches start blooming. Each technique in the field of virtual histology from confocal microscopy to optical coherence tomography have its own advantages and limitations (limited field of view, tissue fixation, photo bleaching, depth penetration in tissues). In most of those modalities, staining of the tissues or contrast agents (such as iodine type with micro-CT) are needed to get visual information about soft tissues (e.g. blood vessels) [5]. In this context, we propose to use synchrotron radiation phase contrast micro-tomography (SR-PCT) to visualize and extract information about the vascular network from unstained mouse liver. Synchrotron radiation, by means of its high brilliance and both spatially and temporal coherency, can give access to the X-ray photon phase information when getting through the sample. As soft tissues have a low absorption coefficient, classic X-ray imaging techniques do not provide an interpretable image of blood vessels without the use of contrast agent such as iodine ones. By taking advantage of the properties previously mentioned, SR-PCT can give access to a whole set of visual information about soft tissues (previous works have been successfully conducted for mouse brain imaging [3]) and especially blood vessels in our case. The technique we are using is called free space propagation and is of practical interest for its simple optical setup and its fast acquisition time. Acquisitions were performed on ID19 beamline at the European Synchrotron Radiation Facility (Grenoble, France). In this specific setup, we are able to acquire a volume of 2560 x 2560 x 1400 pixels representing a field of view of 16.64 x 16.64 x 9.1 mm3 in three minutes with an approximative isotropic spatial resolution of 6.5µm. Thanks to this system, we have acquired four mouse livers under different conditions: three with blood washing and one without, several fixation agents were employed. In this talk, we propose to analyze these large images to provide computational representations of liver blood vessels (see [4] for illustrations). We have first calculated MinIP (Minimum Intensity Projection) images through sets of 100 slices, then binarized to isolate vessels from the rest of soft tissues and the background. The calculation of robust 2D skeleton allows to reconstruct the vascular network from these images afterwards [2]. We have also experimented the calculation of a complete 3D reconstruction, which is not achievable directly due to the high amount of image data to be processed. Hence, the input volumes have been decomposed into groups of 100 slices independently treated with 3D vesselness enhancement algorithms, and results have been grouped back together. To do so, we have tested two different approaches leading to coarse-to-fine representations of the complete hepatic vascular system [1]. As a future prospect, we plan to investigate the use of both 2D and 3D representations in injecting geometrical and topological knowledge in computer analysis of medical imaging (CT or MRI).
References:
[1] Lebre et al. Medical Image Processing and Numerical Simulation for Digital Hepatic Parenchymal Blood Flow. In SASHIMI@MICCAI 2017.
[2] Leborgne and Vacavant. Robust Computations of Reeb Graphs in 2-D Binary Images. In CTIC 2016.
[3] Rositi et al. Fast virtual histology of unstained mouse brains using in-line X-ray phase tomography. In European Molecular Imaging Meeting 2015.
[4] Rositi et al. http://rositi.iut-lepuy.fr/2017/11/20/neubias-submission/
[5] Yoon et al. Three-Dimensional Imaging of Hepatic Sinusoids in Mice Using Synchrotron Radiation Micro-Computed Tomography. PLoS ONE, 8(7): e68600, 2013.
Liver mouse acquisitions:

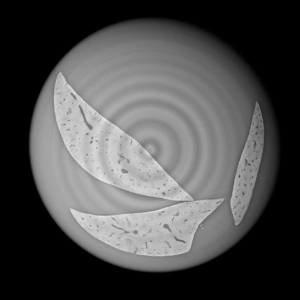
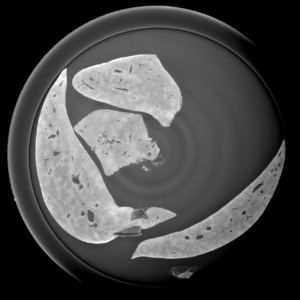
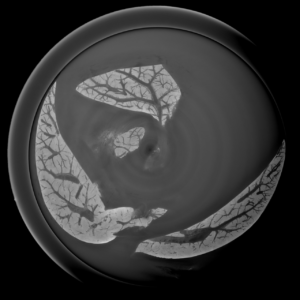
2D analysis and robust skeletonization:

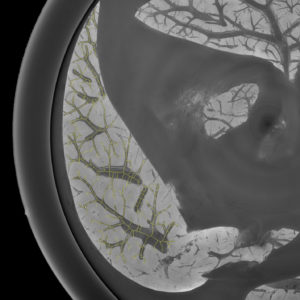
3D analysis and multi-scale reconstruction of blood vessels with two different algorithms: